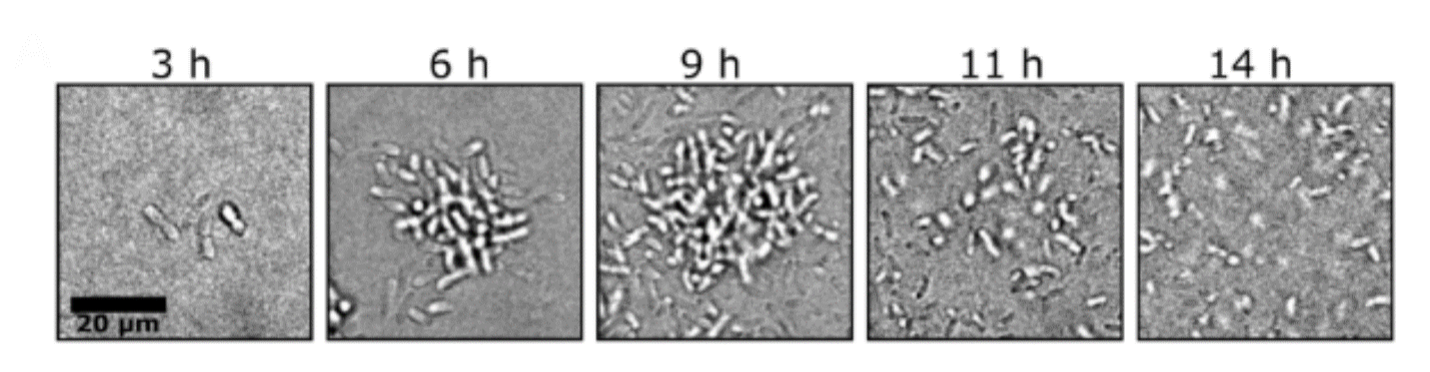
The model pathogen Vibrio cholerae forms biofilms in its aquatic habitat, biofilm cells are especially virulent in mouse models of cholera disease, and biofilms are thought to be critical for cholera transmission. Studies of V. cholerae biofilms have predominantly focused on matrix overproducing strains that constitutively exist in the biofilm mode and that do not disperse. This research strategy has propelled understanding of V. cholerae biofilm attachment and maturation, revealing that the second messenger cyclic diguanylate is a master regulator of biofilm formation, and that expression of vibrio polysaccharide biosynthetic genes are required. The strategy of characterizing constitutive biofilm formers, while successful for uncovering factors that promote biofilm formation, has necessarily precluded studies of biofilm dispersal. Recently, we employed a microscopy assay that allowed us to monitor the full wild-type V. cholerae biofilm lifecycle. We combined this assay with high-content imaging of randomly mutagenized WT V. cholerae to identify genes required for biofilm dispersal. Investigation of the proteins encoded by the genes allowed us to characterize the signaling relays, matrix-digestion enzymes, and motility components required for biofilm dispersal, a key stage in the lifecycle of the global pathogen V. cholerae.

