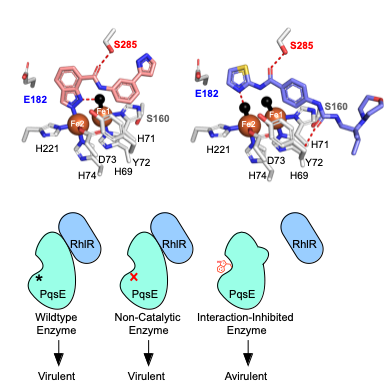
Proteins involved in quorum sensing (QS) present an exciting opportunity for the development of new antibiotics. In the human pathogen Pseudomonas aeruginosa, for instance, QS is essential for driving virulence and establishing infections, but is nonessential for survival. Therefore, antimicrobial agents that target QS mechanisms have the potential to render P. aeruginosa avirulent without spurring resistant mutations. The QS network in P. aeruginosa consists of several interconnected systems and proteins such as autoinducer synthases, receptor/transcription factors, and regulators of unknown function. While many of these proteins are difficult to purify and structurally undefined, one enzyme, PqsE, is an ideal subject for in vitro biochemical assays and is essential for virulence phenotypes in P. aeruginosa. However, the mechanism by which PqsE contributes to QS has been elusive since the discovery that it is not needed for the function of its annotated biosynthetic pathway. Rather, PqsE is known to play a role in the parallel Rhl QS branch.
We have developed a series of tools for studying the functions of PqsE both in vitro and in vivo. These tools include several biochemical assays, small molecule PqsE binders, and mutations in and around the active site of PqsE. A consistent observation that has thwarted efforts to develop a PqsE-targeting antimicrobial agent was that molecules that potently inhibited enzyme activity of PqsE in vitro, never succeeded at inhibiting PqsE-driven virulence in vivo. In order to determine whether targeting PqsE could inhibit virulence, we designed mutations in the active site that mimic the effects of an inhibitor binding in this site. These mutations indeed disabled PqsE enzyme activity, but additionally, they were found to affect PqsE’s ability to form a complex with the QS receptor/transcription factor, RhlR. Through rigorous characterization of various point mutations in PqsE using both in vitro and in vivo assays, we determined that it is the protein-protein interaction with RhlR, and not enzyme activity of PqsE, that is responsible for driving virulence phenotypes. We are interested in further drug discovery efforts against PqsE, now targeting the PqsE-RhlR interaction. Furthermore, we are taking a similar chemical genetic approach to determine the molecular mechanisms involved throughout the whole QS network in P. aeruginosa.

